| |
p53 database
p53 database
Database curation
Download the database
Improve p53 mutation detection and report
p53 mutation and cancer
|
p53 MUTATIONS IN BLADDER CANCER
Bladder Cancer
The most common type of bladder cancer begins in cells lining the inside of the bladder and is called transitional cell carcinoma (TCC). Bladder cancer is the fourth most common type of cancer in men and the ninth most common cancer in women.
Molecular genetic of bladder cancer
The FGFR3, HRAS, RB1, and TP53 genes are associated with bladder cancer.
As with most cancers, the exact causes of bladder cancer are not known; however, many risk factors are associated with this disease. Chief among them are smoking and exposure to industrial chemicals. Mutations in the FGFR3 gene that arise in the bladder are another important risk factor for developing bladder cancer. Similar changes in other genes, such as RB1, HRAS and TP53, may also increase risk. Each of these genes plays a critical role in regulating the cycle of cell division, preventing cells from dividing too rapidly or in an uncontrolled way. Alterations in these genes may help explain why some bladder cancers grow and spread more rapidly than others.
A family history of bladder cancer is a risk factor for the disease, although most genetic changes that are associated with bladder cancer develop in bladder tissue during a person's lifetime, rather than being inherited from a parent. Some people, however, appear to inherit a reduced ability to break down certain chemicals, which makes them more sensitive to the cancer-causing effects of tobacco smoke and certain industrial chemicals
Spectrum of p53 mutations in bladder cancer
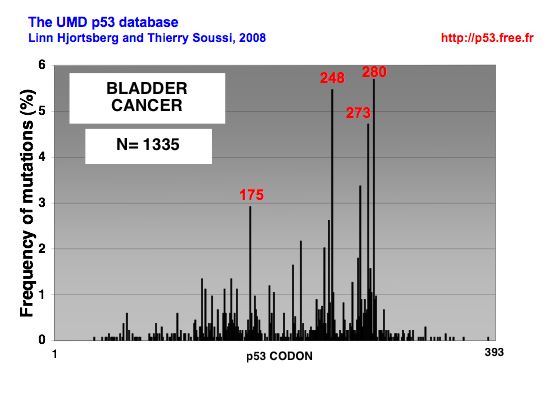 |
DISTRIBUTION OF p53 MUTATIONS IN BLADDER CANCER |
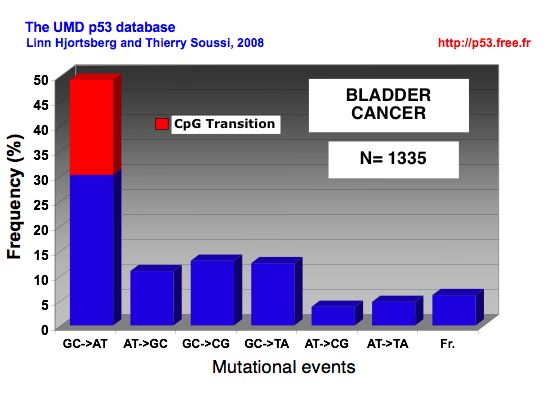 |
MUTATIONAL EVENTS IN BLADDER CANCER |
|
Meta-analysis of p53 loss of function in bladder cancer.
Points ; mean p53 activity as measured by transactivation with the WAF1promoter ; bars , 95 % CI. The mean and 95 % CI of p53 activity for all studies combined for a specific type of cancer is shown on the far left of each graph.
Horizontal line, mean of the combined studies. The publication code is indicated on the x-axis : the first number is an anonymous ID for the publication and the second number indicates the number of p53 mutants included in this study. Studies are presented from left to right in decreasing order of number of p53 mutants. The y-axis corresponds to p53 transactivation activity, with a value of 1.5 for the negative control and a value of 2.5 for 100 % of wild-type activity. Only studies with 5 or more p53 mutations are shown on the graph.
More information about this statistical analysis can be found in this article:
Soussi, T., Asselain, B., Hamroun, D., Kato, S., Ishioka, C., Claustres, M. and Beroud, C. (2006) Meta-analysis of the p53 mutation database for mutant p53 biological activity reveals a methodologic bias in mutation detection. Clin Cancer Res, 12, 62-69. Download the pdf
|
|
|






