| |
p53 database
p53 database
Download the database
Database curation
Improve p53 mutation detection and report
p53 mutation in cell lines
(new)
p53 mutation and cancers: an extensive analysis of p53 mutation in every cancer
The p53 Mutation Handbook
p53 mutation distribution
p53 mutation in breast cancer: about apples, oranges and unusual p53 mutations:
a response to Patoc et al.
|
How to improve p53 mutation detection and reports
Detection
Reports
MUT-P53, an novel excel® spreadsheet to check p53 mutations accuracy before publication
Improving detection of p53 mutation
Strategy
First, only molecular analysis should be performed, as immuno-histochemical analysis cannot distinguish the various types of mutations. It also misses frameshift and nonsense mutation (11.3% and 7.5%, respectively, of mutations found in the p53 mutation database).
Second, TP53 analysis should not be restricted to exons 5–8, as this leads to an unacceptable bias (see figure). Ideally, the entire coding region of TP53 should be analyzed (exons 2–11), including the splice junctions, although analysis of exons 4–10 might be acceptable because it would miss fewer than 1% of all mutations. Richard Iggo and colleagues have developed an assay in yeast that allows the screening of codons 52–364 (68% of exons 4–10) using mRNA as starting material. Ultimately, however, genomic sequencing should be performed, as analysis of RNA can also lead to under-representation of splicesite or nonsense mutations. DNA chip analysis could be one of the favorite methodologies in the future, as it combines good sensitivity with high throughput.
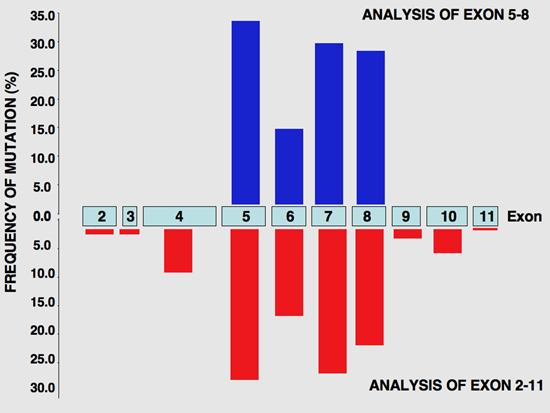 |
Distribution of mutational events in each exon of the TP53 gene. Studies focusing on the central region (exons 5–8,blue bars, top) are compared with those analyzing all coding regions (exons 2–11, red bar, bottom).
.
|
Improving reports
Analysis of the literature reporting TP53 mutations shows that 8% of reports display typographical errors, with a marked increase over recent years. These errors are sometimes isolated, but, in some cases, they concern several or even all mutations described in a single article.
Furthermore, some papers report an unusual profile of p53 mutations, the accuracy of which is difficult to assess. In order to manage these problems, we have developed MUT-TP53, an accurate and powerful tool that will automatically process p53 mutations and generate tables ready for publication that will decrease the risk of typographical errors. Furthermore, using functional and statistical information derived from the UMD p53 database, this matrix can be used to assess the biological activity and likelihood of each TP53 mutant
Download MUT-P53
(you will need Excel from Microsoft to run this matrix)
Soussi T, Rubio-Nevado JM, Ishioka C (2006) MUT-TP53: a versatile matrix for TP53 mutation verification and publication. Hum Mutat 27: 1151-1154.
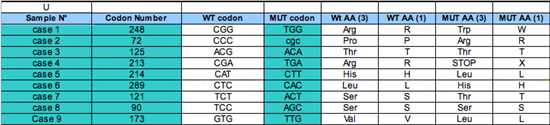 |
Input table
The user only needs to complete column 1 (sample identification), 2 (codon position) and 3 (mutant codon). Other columns, 2 (wt codon), 3 to 5 (wt and mutant amino acids in 1 or 3 letter codes) will be automatically calculated by the macro included in the matrix. |
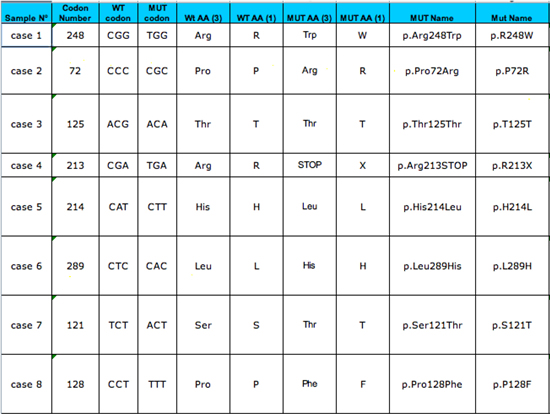 |
Output table (left part)
Once the input table has been completed (up to 150 cases can be included), the output table is automatically generated. It will include the information of the input table plus the mutant name using the international nomenclature. |
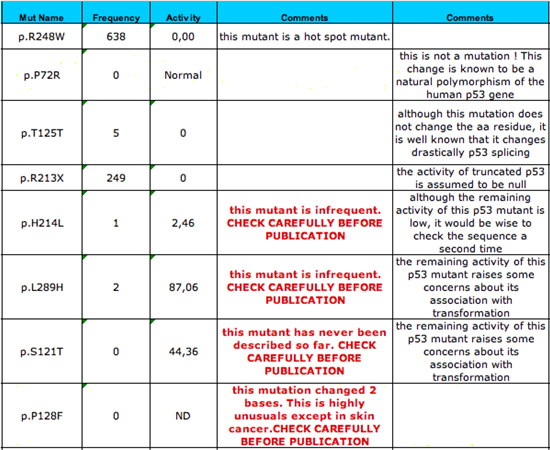 |
Output table (right part)
For each mutant, the table includes its frequency in the database (if available) and the residual activity of the mutant p53 based on the functional activity assay of 3,000 mutants performed by Kato et al.
The figure displays various possibilities. The c.742C>T (p.R248W) TP53 mutant is a well known hot spot mutant that has been found 638 times and it has no residual activity. The finding of this mutant therefore does not raise any questions. The c215C>G (p.R72P) TP53 variant is not a mutant, but, as indicated, a natural polymorphism in exon 4 of the p53 gene. It should not be included in a mutation table except in very specific cases. The c.375G>T (p.T125T) variant seems to be a "neutral" polymorphism. In fact, it has been shown that it very drastically changes p53 splicing. A warning message about this possibility is generated for each mutation that does not change p53 amino acid residues. The c.614A>T (p.H214L) mutant is a very rare variant despite its low remaining activity. A warning message indicates this fact and invites the user to check the mutation. Similarly, a warning message is displayed for the c.866T>A (p.L289H) mutant, which is also very rare and has only a mild effect on p53 activity. For the c.361T>A (p.S121T) mutant, which has never been described to date, a specific warning message is also displayed and the authors should carefully check the experimental data. The c.[382C>T;383C>T] (p.P128F) mutant is due to a mutation that changes two bases in the codon. This is a very unusual observation for the p53 gene except in skin cancer and generally corresponds to a typographical error and these mutants are very infrequent in the database. A specific warning message flags this mutant. |
____________________________________________________________________________________
The UMD p53 database is protected by the European Union Council Directive N° 96/9/EC, OJ (L77) 20 (1996). Extracting, copying or reuse of data from databases without permission are covered by this directive
|
|






