| |
Our Work
p53 in Lung cancer
The Yeast assay
p53 Chips and LDR
|
p53 - Lung cancer and DNA Chips
More information on the the oligonucleotide array: Click here --> p53 Chips and LDR
More information on the Yeast assay (FASAY): Click here -> FASAY
Fouquet, C., Antoine, M., Tisserand, P., Favis, R., Wislez, M., Commo, F., Rabbe, N., Carette, M.F., Milleron, B., Barany, F., Cadranel, J., Zalcman, G. and Soussi, T. (2004) Rapid and sensitive p53 alteration analysis in biopsies from lung cancer patients using a functional assay and a universal oligonucleotide array: a prospective study. Clin Cancer Res, 10, 3479-3489. Download the pdf
Download the supplementary materials
Purpose: Molecular profiling of alterations associated with lung cancer holds the promise to define clinical parameters such as response to treatment or survival. Because less than 5% of small cell lung cancers and less than 30% of non-small cell lung cancers are surgically respectable, molecular analysis will perforce rely on routinely available clinical samples such as biopsies. Identifying tumor mutations in such samples will require a sensitive and robust technology to overcome signal from excess amounts of normal DNA.
Experimental Design : p53 mutation status was assessed from the DNA and RNA of biopsies collected prospectively from 83 patients with lung cancer. Biopsies were obtained either by conventional bronchoscopy or CT-guided percutaneous biopsy. Matched surgical specimens were available for 22 patients. Three assays were used: direct sequencing, a functional assay in yeast, and a newly developed PCR/LDR/ Universal DNA array assay.
Results: Using the functional assay, p53 mutation was found in 62% of biopsies and 64% of surgical specimens with a concordance of 80%. The sensitivity of the functional assay was determined to be 5%. Direct sequencing confirmed mutations in 92% of surgical specimens, but in only 78% of biopsies. The DNA array confirmed 100% of mutations in both biopsies and surgical specimens. Using this newly developed DNA array, we demonstrate the feasibility of directly identifying p53 mutations in clinical samples containing less than 5% of tumor cells.
Conclusion: The versatility and sensitivity of this new array assay should allow further development of mutation profiling arrays that could be applied to biological samples with a low tumor cell content, such as bronchial aspirates, bronchoalveolar lavage fluid or serum.
70 patients have been fully analyzed.
RNA and DNA have been successfully extracted for all the clinical specimen (biopsies and surgical specimen). In every case, the RNA was suitable for RT-PCR and FASAY. p53 mutation was identified in 8 samples. The correspondance of p53 mutations in biopsies and surgical specimen with the FASAY was 100%.
Genomic DNA sequencing has led to the confirmation of 80% of these mutations confirming the low senstitivity of direct sequencing when normal DNA contamination is too high in the clinical sample. With the FASAY, we are able to detect p53 mutation in sample with 15% of tumoral cells. Such sensitivity cannot be reach by automatic DNA sequencing.
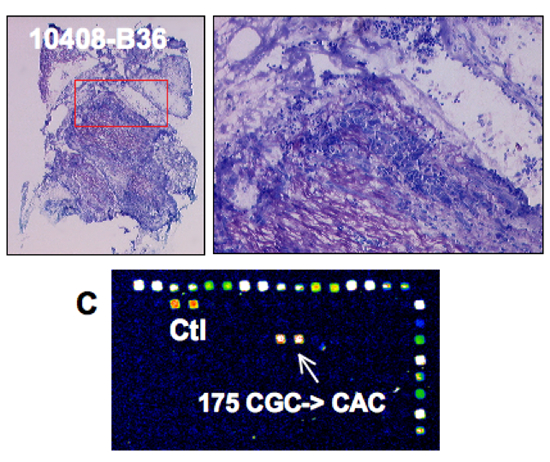 |
p53 analysis in a surgical specimen
p53 mutation detection:
Direct sequencing: YES
FASAY: YES
DNA chip: YES |
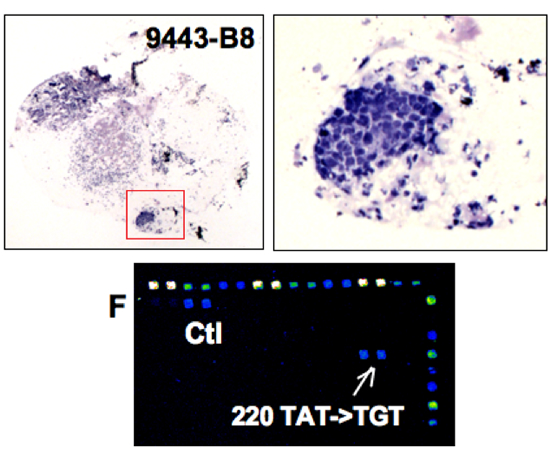 |
p53 analysis in a biopsy
p53 mutation detection:
Direct sequencing: NO
FASAY: YES
DNA chip: YES |
 |
p53 analysis in a biopsy
p53 mutation detection:
Direct sequencing: NO
FASAY: YES
DNA chip: YES |
|
|






