| |
p53 Information
p53 information
p53 story
p53 monoclonal antibodies
p53 pathways
p53 gene
mdm family
mouse models
|
p53 mouse p53 R172H (R175H in human)
Go back to the list
Liu, G., T. J. McDonnell, R. Montes de Oca Luna, M. Kapoor, B. Mims, A. K. El-Naggar, and G. Lozano (2000) Proc Natl Acad Sci U S A 97:4174-4179.
High metastatic potential in mice inheriting a targeted p53 missense mutation
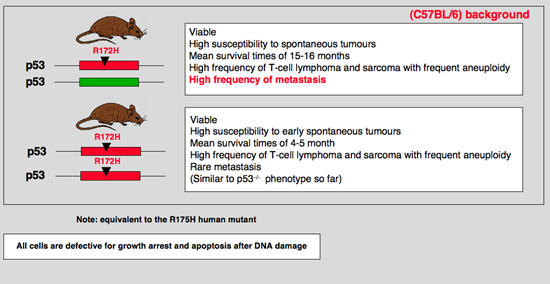
To understand the relevance of p53 missense mutations in vivo, we generated a mouse containing an arg-to-his substitution at p53 amino acid 172, which corresponds to the R175H hot-spot mutation in human tumors by homologous recombination. Inadvertently, this mouse contains the additional deletion of a G nucleotide at a splice junction that attenuates levels of mutant p53 to near wild-type levels. Mice heterozygous for the mutant allele differed from p53(+/-) mice in tumor spectrum, with a significant increase in the number of carcinomas and a slight decrease in the number of lymphomas. More importantly, the osteosarcomas and carcinomas that developed in these mutant mice frequently metastasized (69% and 40%, respectively). In contrast, metastasis is rare in osteosarcomas of p53(+/-) mice. Loss of heterozygosity studies of tumors indicated loss of heterozygosity in only 1 of 11 tumors. These data indicate clear differences between a p53 missense mutation and a null allele in tumorigenesis in vivo and suggest that the p53R172HDeltag mutant represents a gain-of-function allele. |
 |
Lang, G. A., T. Iwakuma, Y. A. Suh, G. Liu, V. A. Rao, J. M. Parant, Y. A. Valentin-Vega, T. Terzian, L. C. Caldwell, L. C. Strong, A. K. El-Naggar, and G. Lozano (2004) Cell 119:861-872.
Gain of Function of a p53 Hot Spot Mutation in a Mouse Model of Li-Fraumeni Syndrome
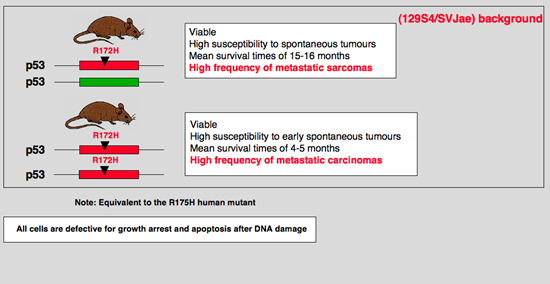
Individuals with Li-Fraumeni syndrome carry inherited mutations in the p53 tumor suppressor gene and are predisposed to tumor development. To examine the mechanistic nature of these p53 missense mutations, we generated mice harboring a G-to-A substitution at nucleotide 515 of p53 (p53(+/515A)) corresponding to the p53R175H hot spot mutation in human cancers. Although p53(+/515A) mice display a similar tumor spectrum and survival curve as p53(+/-) mice, tumors from p53(+/515A) mice metastasized with high frequency. Correspondingly, the embryonic fibroblasts from the p53(515A/515A) mutant mice displayed enhanced cell proliferation, DNA synthesis, and transformation potential. The disruption of p63 and p73 in p53(-/-) cells increased transformation capacity and reinitiated DNA synthesis to levels observed in p53(515A/515A) cells. Additionally, p63 and p73 were functionally inactivated in p53(515A) cells. These results provide in vivo validation for the gain-of-function properties of certain p53 missense mutations and suggest a mechanistic basis for these phenotypes. |
 |
Olive, K. P., D. A. Tuveson, Z. C. Ruhe, B. Yin, N. A. Willis, R. T. Bronson, D. Crowley, and T. Jacks (2004) Cell 119:847-860.
Mutant p53 Gain of Function in Two Mouse Models of Li-Fraumeni Syndrome
The p53 tumor suppressor gene is commonly altered in human tumors, predominantly through missense mutations that result in accumulation of mutant p53 protein. These mutations may confer dominant-negative or gain-of-function properties to p53. To ascertain the physiological effects of p53 point mutation, the structural mutant p53(R172H) and the contact mutant p53(R270H) (codons 175 and 273 in humans) were engineered into the endogenous p53 locus in mice. p53(R270H/+) and p53(R172H/+) mice are models of Li-Fraumeni Syndrome; they developed allele-specific tumor spectra distinct from p53(+/-) mice. In addition, p53(R270H/)(-) and p53(R172H/)(-) mice developed novel tumors compared to p53(-/-) mice, including a variety of carcinomas and more frequent endothelial tumors. Dominant effects that varied by allele and function were observed in primary cells derived from p53(R270H/+) and p53(R172H/+) mice. These results demonstrate that point mutant p53 alleles expressed under physiological control have enhanced oncogenic potential beyond the simple loss of p53 function. |
Go back to the list
|
|






